news model
license











































































































| ~ | 8528 (T/C) | 8528 (T/G) | 8528 (T/A) | 8528 (T/C) |
|---|---|---|---|---|
| ~ | 8528 (TGA/CGA) | 8528 (TGA/GGA) | 8528 (ATG/AAG) | 8528 (ATG/ACG) |
| MitImpact id | MI.1789 | MI.1788 | MI.3 | MI.4 |
| Chr | chrM | chrM | chrM | chrM |
| Start | 8528 | 8528 | 8528 | 8528 |
| Ref | T | T | T | T |
| Alt | C | G | A | C |
| Gene symbol | MT-ATP8 | MT-ATP8 | MT-ATP6 | MT-ATP6 |
| Extended annotation | mitochondrially encoded ATP synthase membrane subunit 8 | mitochondrially encoded ATP synthase membrane subunit 8 | mitochondrially encoded ATP synthase membrane subunit 6 | mitochondrially encoded ATP synthase membrane subunit 6 |
| Gene position | 163 | 163 | 2 | 2 |
| Gene start | 8366 | 8366 | 8527 | 8527 |
| Gene end | 8572 | 8572 | 9207 | 9207 |
| Gene strand | + | + | + | + |
| Codon substitution | TGA/CGA | TGA/GGA | ATG/AAG | ATG/ACG |
| AA position | 55 | 55 | 1 | 1 |
| AA ref | W | W | M | M |
| AA alt | R | G | K | T |
| Functional effect general | missense | missense | start_lost | start_lost |
| Functional effect detailed | missense | missense | start_lost | start_lost |
| OMIM id | 516070 | 516070 | 516060 | 516060 |
| HGVS | NC_012920.1:g.8528T>C | NC_012920.1:g.8528T>G | NC_012920.1:g.8528T>A | NC_012920.1:g.8528T>C |
| HGNC id | 7415 | 7415 | 7414 | 7414 |
| Respiratory Chain complex | V | V | V | V |
| Ensembl gene id | ENSG00000228253 | ENSG00000228253 | ENSG00000198899 | ENSG00000198899 |
| Ensembl transcript id | ENST00000361851 | ENST00000361851 | ENST00000361899 | ENST00000361899 |
| Ensembl protein id | ENSP00000355265 | ENSP00000355265 | ENSP00000354632 | ENSP00000354632 |
| Uniprot id | P03928 | P03928 | P00846 | P00846 |
| Uniprot name | ATP8_HUMAN | ATP8_HUMAN | ATP6_HUMAN | ATP6_HUMAN |
| Ncbi gene id | 4509 | 4509 | 4508 | 4508 |
| Ncbi protein id | YP_003024030.1 | YP_003024030.1 | YP_003024031.1 | YP_003024031.1 |
| PhyloP 100V | 7.698 | 7.698 | 7.698 | 7.698 |
| PhyloP 470Way | 0.742 | 0.742 | 0.742 | 0.742 |
| PhastCons 100V | 1 | 1 | 1 | 1 |
| PhastCons 470Way | 0.965 | 0.965 | 0.965 | 0.965 |
| PolyPhen2 | probably_damaging | probably_damaging | possibly_damaging | possibly_damaging |
| PolyPhen2 score | 1.0 | 0.98 | 0.8 | 0.8 |
| SIFT | neutral | neutral | deleterious | deleterious |
| SIFT score | 0.31 | 0.34 | 0 | 0 |
| SIFT4G | Damaging | Damaging | Damaging | Damaging |
| SIFT4G score | 0.0 | 0.0 | 0.0 | 0.0 |
| VEST | Neutral | Neutral | Neutral | Neutral |
| VEST pvalue | 0.15055088 | 0.13200123 | 0.25 | 0.47 |
| VEST FDR | 0.85 | 0.85 | 0.65 | 0.65 |
| Mitoclass.1 | damaging | damaging | damaging | damaging |
| SNPDryad | Pathogenic | Pathogenic | Pathogenic | Pathogenic |
| SNPDryad score | 0.97 | 0.93 | 0.99 | 0.97 |
| MutationTaster | . | . | Disease | Disease automatic |
| MutationTaster score | . | . | 1 | 1 |
| MutationTaster converted rankscore | . | . | 0.81001 | 0.81001 |
| MutationTaster model | . | . | complex_aae | complex_aae |
| MutationTaster AAE | . | . | N2T | N2T |
| fathmm | . | . | Tolerated | Tolerated |
| fathmm score | . | . | 4.64 | 4.63 |
| fathmm converted rankscore | . | . | 0.01799 | 0.01817 |
| AlphaMissense | likely_pathogenic | likely_benign | . | . |
| AlphaMissense score | 0.7923 | 0.3215 | . | . |
| CADD | Deleterious | Deleterious | Deleterious | Deleterious |
| CADD score | 3.65294 | 3.958969 | 6.259636 | 3.65294 |
| CADD phred | 23.2 | 23.6 | 28.9 | 23.2 |
| PROVEAN | Damaging | Damaging | Damaging | Damaging |
| PROVEAN score | -12.74 | -12.02 | -4.89 | -4.97 |
| MutationAssessor | . | medium | . | . |
| MutationAssessor score | . | 3.38 | . | . |
| EFIN SP | Neutral | Neutral | Neutral | Neutral |
| EFIN SP score | 0.85 | 0.84 | 0.614 | 0.704 |
| EFIN HD | Damaging | Damaging | Damaging | Damaging |
| EFIN HD score | 0.06 | 0.104 | 0.09 | 0.126 |
| MLC | Deleterious | Deleterious | Deleterious | Deleterious |
| MLC score | 0.7 | 0.7 | 0.7 | 0.7 |
| PANTHER score | . | . | . | . |
| PhD-SNP score | . | . | . | . |
| APOGEE1 | Pathogenic | Pathogenic | Pathogenic | Pathogenic |
| APOGEE1 score | 0.58 | 0.64 | 0.58 | 0.63 |
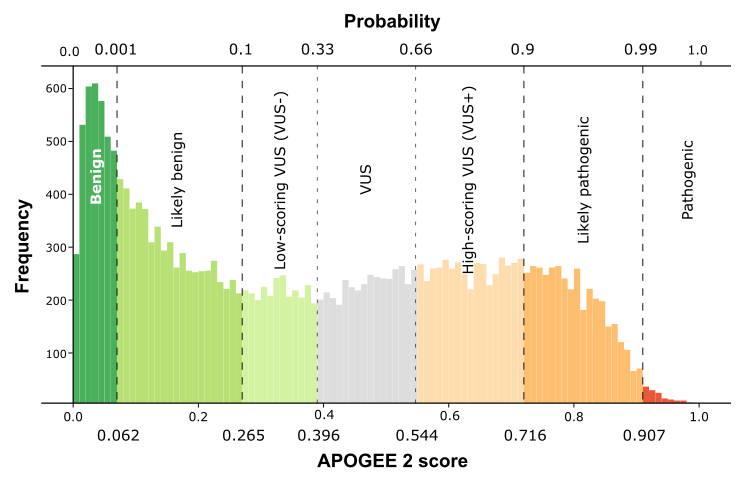
| APOGEE2 | Likely-pathogenic | Likely-pathogenic | VUS+ | Likely-pathogenic |
| APOGEE2 score | 0.841959764587885 | 0.820374350074967 | 0.565745889740842 | 0.759827033070099 |
| CAROL | deleterious | deleterious | deleterious | deleterious |
| CAROL score | 1 | 0.98 | 1 | 1 |
| Condel | neutral | neutral | neutral | neutral |
| Condel score | 0.16 | 0.18 | 0.1 | 0.1 |
| COVEC WMV | deleterious | deleterious | deleterious | deleterious |
| COVEC WMV score | 2 | 2 | 3 | 3 |
| MtoolBox | deleterious | deleterious | deleterious | deleterious |
| MtoolBox DS | 0.9 | 0.87 | 0.73 | 0.76 |
| DEOGEN2 | . | . | Tolerated | Tolerated |
| DEOGEN2 score | . | . | 0.238148 | 0.311115 |
| DEOGEN2 converted rankscore | . | . | 0.60599 | 0.68302 |
| Meta-SNP | . | . | . | . |
| Meta-SNP score | . | . | . | . |
| PolyPhen2 transf | low impact | low impact | . | . |
| PolyPhen2 transf score | -3.6 | -2.36 | . | . |
| SIFT_transf | medium impact | medium impact | . | . |
| SIFT transf score | 0.1 | 0.13 | . | . |
| MutationAssessor transf | high impact | high impact | . | . |
| MutationAssessor transf score | 2.07 | 2.07 | . | . |
| CHASM | Neutral | Neutral | Neutral | Neutral |
| CHASM pvalue | 0.08 | 0.06 | 0.42 | 0.24 |
| CHASM FDR | 0.85 | 0.85 | 0.9 | 0.9 |
| ClinVar id | 9640.0 | . | . | 9640.0 |
| ClinVar Allele id | 24679.0 | . | . | 24679.0 |
| ClinVar CLNDISDB | MONDO:MONDO:0044970,MedGen:C0751651,Orphanet:68380|Human_Phenotype_Ontology:HP:0005152,MONDO:MONDO:0010771,MedGen:C1708371,OMIM:500000,Orphanet:137675|MONDO:MONDO:0010777,MedGen:C2748884,OMIM:500006 | . | . | MONDO:MONDO:0044970,MedGen:C0751651,Orphanet:68380|Human_Phenotype_Ontology:HP:0005152,MONDO:MONDO:0010771,MedGen:C1708371,OMIM:500000,Orphanet:137675|MONDO:MONDO:0010777,MedGen:C2748884,OMIM:500006 |
| ClinVar CLNDN | Mitochondrial_disease|Histiocytoid_cardiomyopathy|Cardiomyopathy,_infantile_hypertrophic | . | . | Mitochondrial_disease|Histiocytoid_cardiomyopathy|Cardiomyopathy,_infantile_hypertrophic |
| ClinVar CLNSIG | Likely_pathogenic | . | . | Likely_pathogenic |
| MITOMAP Disease Clinical info | Infantile cardiomyopathy / hyperammonemia | . | . | Infantile cardiomyopathy / hyperammonemia |
| MITOMAP Disease Status | Cfrm [LP] | . | . | Cfrm [LP] |
| MITOMAP Disease Hom/Het | +/+ | ./. | ./. | +/+ |
| MITOMAP General GenBank Freq | 0.0% | . | . | 0.0% |
| MITOMAP General GenBank Seqs | 0 | . | . | 0 |
| MITOMAP General Curated refs | 26803244;30642647;19188198;27409572;26741492;34298071;33180048;30763462;21457906 | . | . | 26803244;30642647;19188198;27409572;26741492;34298071;33180048;30763462;21457906 |
| MITOMAP Variant Class | disease | . | . | disease |
| gnomAD 3.1 AN | . | . | . | . |
| gnomAD 3.1 AC Homo | . | . | . | . |
| gnomAD 3.1 AF Hom | . | . | . | . |
| gnomAD 3.1 AC Het | . | . | . | . |
| gnomAD 3.1 AF Het | . | . | . | . |
| gnomAD 3.1 filter | . | . | . | . |
| HelixMTdb AC Hom | . | . | . | . |
| HelixMTdb AF Hom | . | . | . | . |
| HelixMTdb AC Het | . | . | . | . |
| HelixMTdb AF Het | . | . | . | . |
| HelixMTdb mean ARF | . | . | . | . |
| HelixMTdb max ARF | . | . | . | . |
| ToMMo 54KJPN AC | . | . | . | . |
| ToMMo 54KJPN AF | . | . | . | . |
| ToMMo 54KJPN AN | . | . | . | . |
| COSMIC 90 | . | . | . | . |
| dbSNP 156 id | . | . | . | rs387906422 |